What is the evolutionary significance of introns?
September 10, 2009

- Related Topics:
- DNA basics,
- RNA,
- Noncoding DNA,
- Genes to proteins
A curious adult from Northern Ireland asks:
“I would really appreciate your expertise and your notions as to the evolutionary significance of introns. What do they do in eukaryotic genomes and why do prokaryotes not have introns?”
This is a great question! Scientists are still trying to figure out why some eukaryotes like yeast have only a few introns while others like humans have tens of thousands of them. And why prokaryotes don’t have any spliceosomal introns*.
There are actually two competing theories. One is called introns-early (IE). It says that introns used to be in both prokaryotes and eukaryotes, but bacteria and other prokaryotes have since lost them.
The other theory, as you might have guessed, is called introns-late (IL). It says that introns developed in eukaryotes after they split from prokaryotes.
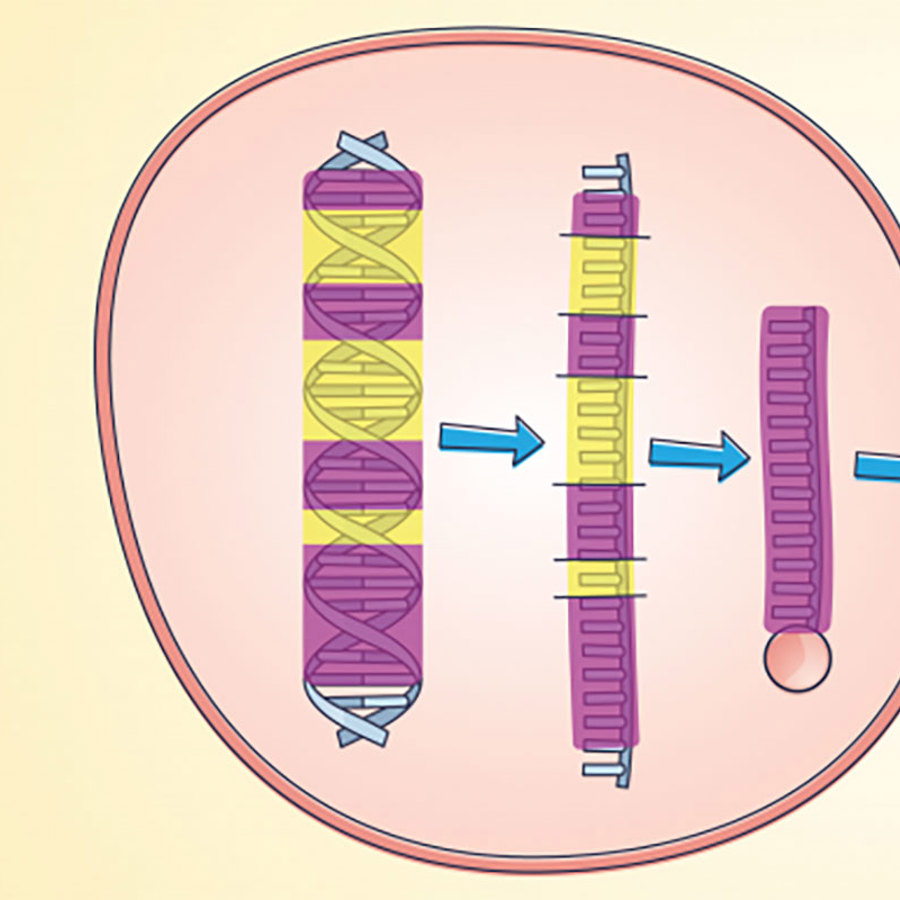
Without a time machine, it is obviously hard to tell which of these models is right. I’ll go into more detail about evidence for both models later in this article. Scientists are looking at a lot of DNA from many different beasts to look for clues about where introns came from.
Before talking about any results, we need to make sure we're all on the same page. What I’ll do first is briefly go over what we mean by genes, genomes, introns, etc. Then we can delve a bit deeper into some of the ideas about why we have these pesky introns at all.
(*Prokaryotes actually do have a certain type of introns. The RNA produced from these introns have their own special activity – they can splice themselves. Some scientists believe that the eukaryotic introns evolved from these prokaryotic ones.)
Genes, Introns and Proteins, Oh My!
As you probably already know, DNA has the instructions for making and running a living thing. And each type of living thing has a different set of DNA instructions. So, for example, a dog has the instructions to make four legs and an octopus has instructions for making eight.
Many of these instructions are found in long stretches of DNA called genes. Each gene has the instructions for making a particular protein that does a specific job.
Some proteins carry oxygen in our blood. Others digest starch in our food. Or still others make pigments that color our eyes, hair, and skin. And so on. Basically proteins do most of the work of making and running each of us.
Introns are actually big pieces of DNA stuck in the middle of genes. They contain a lot of information and instructions on how specific proteins should be created.
But if the cell doesn’t cut the introns out, it’ll make mostly garbled proteins. Luckily, cells can remove them before they read the gene’s instructions. These introns are cut out pretty early on, at the RNA stage.
Introns and Exons: Cutting and Pasting
Our cells don't go directly from the DNA instructions in genes to proteins. First, they copy those instructions into something called RNA. This RNA (called messenger or mRNA) is then translated into a specific protein.
Introns are chopped out as RNA is made from the DNA. The remaining pieces of protein assembly instructions (called exons) are then pasted together.
Splicing can sometimes happen at different spots along an RNA strand. What then happens is that different bits of RNA get pasted together to make different proteins. Because of splicing, you can get more than one protein from a single gene. These different proteins are called splice variants.
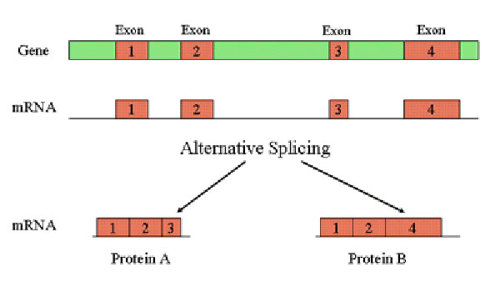
Let’s look at how this all works using the word ‘magnificent.’ We’ll pretend that ‘magnificent’ is a gene.
You can remove various chunks of letters (introns) and get many different words (splice variants). A few words you get are ‘magic,’ ‘nice,’ or ‘age.’ Even though they came from the same place, these words all have different meanings and different lengths.
Now imagine you need to write the sentence, “Harry Potter is magic.” Now imagine that you spliced ‘magnificent’ wrong and ended up with ‘nice’ instead of ‘magic.’ You get, “Harry Potter is nice.” It still makes sense but has a different meaning. And ‘age’ would make for a nonsense sentence!
The same thing can happen with the splicing of mRNA. Depending on where an mRNA is cut and pasted, you can end up with different proteins that do different things. Like Harry being magic in one case and nice in the other.
And sometimes splicing can happen in the wrong place. Then you can end up with a protein that can stop working or that gains a new function. These splice variants can have serious consequences such as cancer or disease.
Which is why getting splicing right is so important. Given all of this, introns better have some critical role or it would be easier just not to have them around at all.
Evolutionary Significance of Introns
As I said at the beginning, the number of introns varies greatly between species from less than a hundred to hundreds of thousands per cell. The size of each intron also varies greatly. In simpler organisms they can be quite short, while in higher eukaryotes, they can be very long.
So where did introns come from? Remember there are two competing hypotheses: introns-early (IE) and introns-late (IL). Although we don't know which (if either) happened, I'll explain the basics of these two models and give some evidence for each.
The IE hypothesis proposes that introns are very old and were found in both prokaryote and eukaryote ancestors. The idea is that our earliest ancestors had many very small protein units. Introns then allowed the mixing and matching of these instructions which caused a rapid expansion in the number of genes.
Over time, introns were lost from prokaryotes as a way to make proteins more efficiently. This matters in the hurly burly world of bacteria where growing as fast as possible is often critical for survival.
In the IL hypothesis, introns appeared after the prokaryotes and eukaryotes went their separate ways. So instead of introns being lost from prokaryotes, they were gained in eukaryotes.
Is there an advantage for introns and exons? Remember, most eukaryotes are multicellular organisms. The mixing and matching of exons from the same gene can lead to proteins with different functions.
Eukaryotes might need this diversity in proteins because they have many types of cells all with the same set of genes. Therefore, introns are a really efficient way to generate many different proteins or different amounts of proteins that are unique to a cell type.
Introns might also allow for faster evolution. For example, cutting and pasting exons from existing genes can also create new genes.
Introns Early or Late? Maybe sometime in-between.
So scientists can come up for a rationale for either model. To try to figure out which one is right, scientists are comparing where introns are located in different organisms.
If introns were lost from prokaryotes (IE theory), then we might predict that introns in eukaryotes should be in around the same place as in prokaryotes. At least for some of the oldest introns.
This is because eukaryotes had introns long ago, before splitting with prokaryotes. And eukaryotes evolving from these early ancestors should keep some of these same introns.
If introns were gained in eukaryotes (IL theory), then introns between distant species should be in more random places. This is because as eukaryotes evolved, the species with a higher number of introns should have them inserted in more and different places in their DNA. (This is assuming that introns insert randomly in DNA. That is another story!)
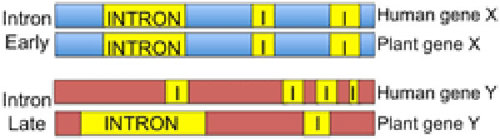
The actual comparison of intron position shows something in-between, where not everything is similar and not everything is random. For example, 25% of human introns are at the exact same position as plant introns1. But 20-68% of introns are specific to that species1.
So, as you can tell, there's still a lot of work scientists need to do to answer your question. Figuring out the DNA sequences of more species and doing more comparisons of closely related species might help solve the mystery of intron evolution.
Read More:
- Roy et al. “The evolution of spliceosomal introns: patterns, puzzles and progress.” Nature Reviews Genetics. (2006)
- “The History of the Intron--Balancing Gains and Losses.” PLoS Biol.(2004)

Author: Gwen Liu
When this answer was published in 2009, Gwen was a Ph.D. candidate in the Department of Microbiology & Immunology, studying the structure and function of microRNA genes in Chang-Zhen Chen’s laboratory. Gwen wrote this answer while participating in the Stanford at The Tech program.
 Skip Navigation
Skip Navigation
