What is the T haplogroup of the paternal Y-DNA?
September 30, 2016

- Related Topics:
- Y chromosome,
- Ancestry,
- Genetic variation,
- Mutation,
- Population genetics
A curious adult from the US asks:
"I just had a DNA test done and it came back that my paternal Y-DNA was the rare haplogroup T. Can you tell me more about the T haplogroup?"
First of all, let me congratulate you on being in good company! Thomas Jefferson is also part of haplogroup T!
Haplogroups are just a way of using DNA to figure out where our ancestors came from. People with common ancestors will share some DNA with each other that they will not share with anyone else. So people in the same haplogroup share some of the same DNA which means they share a common ancestor.
Your question is about your Y-DNA haplogroup, which involves Y chromosome DNA. This DNA traces back along the paternal line. From son to father (and his brothers and their sons) to grandfather (and his brothers and their sons and grandsons) and so on.
The other haplogroup type involves mitochondrial DNA (mtDNA). This is passed from mother to children and so only traces back mom’s female side of the family. Mom and her sisters and their children, your mom’s mom and her sisters and their children and grandchildren and so on.
Y-DNA Haplogroup T
There are 20 main Y-DNA haplogroups, but many of them have subgroups. Technically, any time two people differ at just one tiny spot in their DNA, they could be said to have different haplogroups.
But since that would mean keeping track of millions of groups, scientists instead split us into fewer categories. They only look at differences that are useful – such as those that help us track the migration of early humans.
Paternal Y-DNA doesn’t recombine (or shuffle around) with each generation, meaning we can use it to trace generations back tens of thousands of years!
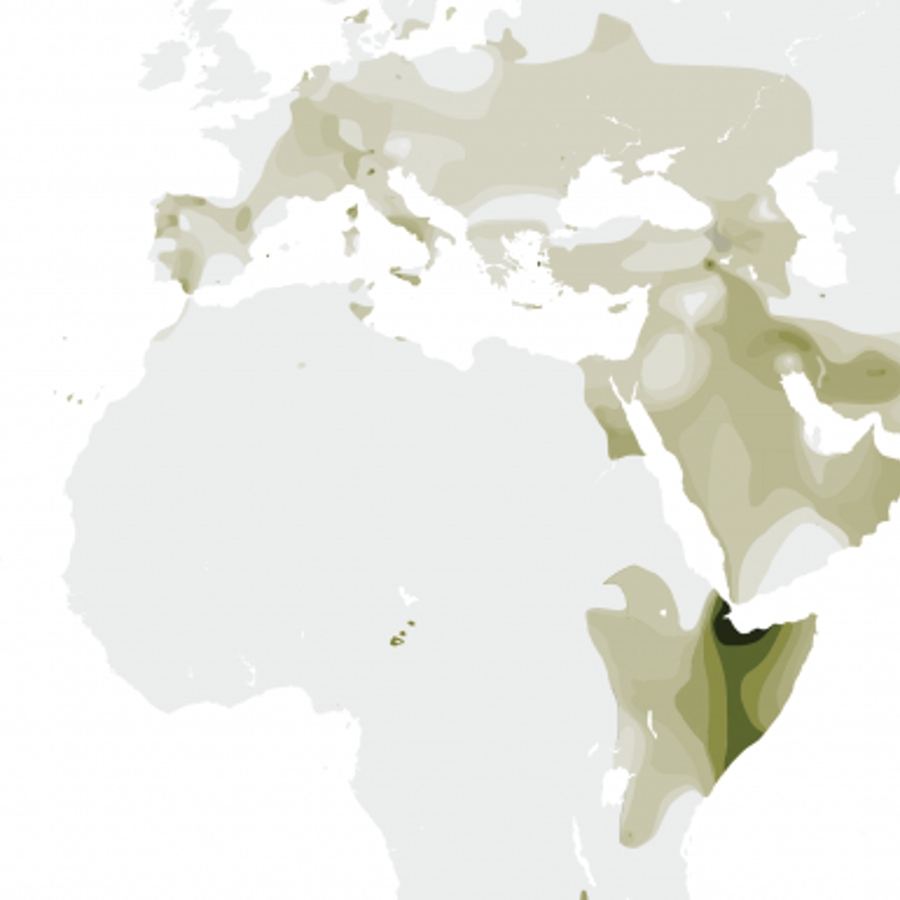
The Y-DNA Haplogroup T is considered unusual because it’s both pretty rare and geographically widespread. Usually you just have one or the other.
Scientists think this group appeared (more on that later) between 15,000 and 22,000 years ago. This is pretty recent as far as human history goes.
Only about 1% of all the people living in Europe are part of this group, but in some areas, this percentage is greater. In Greece and Italy it’s 4%, and in Ibiza, it’s 15%!
Today, the highest concentration of men in the Y-DNA haplogroup live in East Africa and the Middle East. Within the nomadic Fulani people of Cameroon, 18% of males have the DNA of the T Haplogroup!
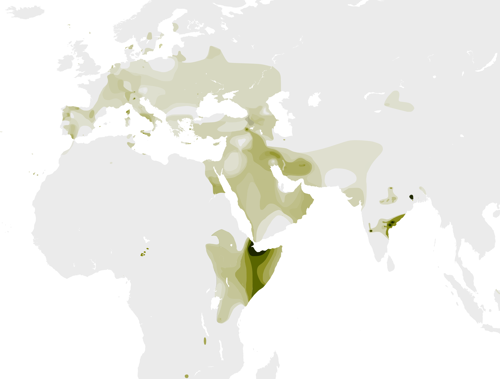
Though right now T Haplogroup males are most common in East Africa, anthropologists (scientists who study the origins of people and civilizations) think it originated in the Fertile Crescent. The Fertile Crescent is a half-moon shaped area in Western Asia containing the areas around the Nile River.
These areas are moist and fertile, unlike the dry lands around them. Because of this hugely important water source, and the fact that there were plenty of edible plants that could be farmed, some of the earliest civilizations flourished in this area.
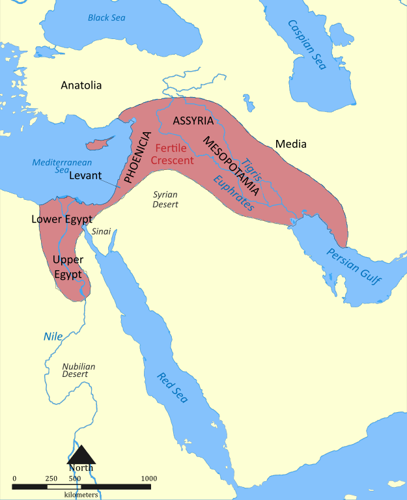
Over long periods of time, groups of these civilizations moved towards modern-day Africa and Europe. And of course they took their Y-DNA with them, establishing new hotspots for the T haplogroup!
That’s how separate areas in Europe and Africa ended up having men of the T haplogroup around. And, how, after many generations, you and Thomas Jefferson ended up being in the same group!
So that’s the T haplogroup. If you want to dig deeper and take a closer look at our DNA differences and why the Y chromosome is so important for haplogroups, read on!
Grouping Different Versions of DNA into Haplogroups
We’ve established that a haplogroup is a group of people who all have a very similar combination of DNA sequences. But let’s see exactly what that means.
Let’s say that a specific bit of DNA on one of my chromosomes looks like this:
ATTCGTCGCGTACTAATTCCCGTGT
This same spot on your chromosome looks like this:
ATTCGTCGCTTACTAATTCCCGTGT
Where I have a G, you have a T instead. Sometime in history, there was a mutation. A mutation is a permanent change in a DNA sequence.
Within our cells, DNA damage can happen as often as 10,000 times per cell per day![1,2] Left unfixed, this damage can lead to mutations.
Luckily our cells are very good at repairing their DNA. But they aren’t perfect. Some slip through and become mutations.
On top of these “spontaneous” mutations, our DNA can also be damaged from outside sources like chemicals or UV light from the sun. Again, left unrepaired, this damage goes on to become mutations.
Of course, in order for mutations to be passed down from parents to children, the DNA of the sperm or egg needs to be mutated. Recent studies found that each newborn has around 60 new mutations.
Now, most of these new mutations that we get aren’t bad. Since DNA is really like our instruction manual, it shouldn’t be too surprising that there are little differences between my DNA and yours. After all, we aren’t all exactly alike!
Now, look again at our sequences again:
ATTCGTCGCGTACTAATTCCCGTGT
ATTCGTCGCTTACTAATTCCCGTGT
Without looking at many other versions of this specific bit of DNA, it’s hard to say if the red letter started as a G, and a mutation happened to change it to a T, or the other way around. Or, a third possibility is that it used to be an A, and two different mutations happened to get to our sequences above.
But what we can say for sure is that our versions of this piece of DNA are different at this position. Each haplogroup has specific changes like this that define this category.
So if we looked at the same areas of our DNAs and found many differences like this, we would likely be in different haplogroups.
It isn’t a huge surprise, then, that people who live in different areas of the world tend to belong to distinct haplogroups. The DNA of people from China happened to mutate in different spots compared to the people of the Netherlands. We can look and see these differences in someone’s DNA and get a good idea about where they came from.
And of course if a group from the Netherlands were to move to, say, Indonesia, then they would take their DNA with them. When scientists looked at the DNA of some of these folks hundreds of years later, they’d see Dutch DNA.
They would know, just by looking at their DNA that at some point a group of people migrated from the Netherlands to Indonesia! This kind of pattern-finding has made it possible for scientists to trace the first humans as they migrated from Africa to Asia and Europe and populated the world.
Why Y-DNA and mtDNA
So simple right? Just see which bunches of DNA differences travel together and you can see who is related to whom.
Well, it turns out that we can’t easily do this for most of our DNA. See, most of our DNA gets shuffled around when it gets passed from parent to child. This can break up these bunches of differences we want to look at and make them hard to trace.
Because of this, we can only trace most of our DNA back five or so generations. There are two exceptions: the Y chromosome (Y-DNA) and mitochondrial DNA (mtDNA).
Y-DNA and mtDNA don’t recombine (or shuffle around) with each generation, meaning we can go way back in time with these. In fact, we can trace generations back tens of thousands of years!
But they do have their limitations. The Y is only passed from father to sons. This means we can only trace back son to father to grandfather and so on. Your mom’s side of the family is invisible.
The mtDNA is passed only from mothers to children. So you can only trace the maternal line back with this, making your dad’s side of the family invisible.
Even with these downsides, scientists have been able to trace back mtDNA and Y-DNA haplogroups tens of thousands of years. Because of these convenient groups, we are closer to understanding where early humans first lived, and how they moved around the world to populate all the different continents. And what’s really cool, is that we can look at the DNA of someone living today, and trace their ancestry way back in time, just like you did by testing your DNA!

Author: Sandra Cristea
When this answer was published in 2016, Sandra was a Ph.D. candidate in the Cancer Biology program working in Julien Sage’s lab, where she studied how to leverage enzymes to better understand and treat small cell lung cancer. Sandra wrote this answer while participating in the Stanford at The Tech program.
 Skip Navigation
Skip Navigation
